Assembling the Cliveome
We recently participated in a collaborative effort to sequence, assemble, and analyze a human genome (GM12878) using the Oxford Nanopore MinION (Jain et al. 2017). As part of that project, Josh Quick and Nick Loman developed a nanopore sequencing protocol capable of generating “ultra-long” reads of length 100 kb and greater. In the paper we predict that reads of such length could enable the most continuous human assemblies to date, with NG50 contig sizes exceeding 30 Mbp. Thus far, we have only collected 5x coverage using the ultra-read protocol and cannot fully test this prediction. However, another human dataset, the “Cliveome”, lets us compare the effect of read length and coverage on nanopore assembly. Here we present a brief analysis of that assembly, which achieved a remarkable contig NG50 of 24.5 Mbp.
The recent Jain et al. preprint includes a model we developed to predict human assembly continuity based on the length and accuracy characteristics of the sequencing reads. Here is an adaptation of Figure 5A from the paper:
A model of expected NG50 contig size when human repeats of a certain length and identity can be correctly resolved. Repeats can be resolved either by long reads that completely span the repeat, or by accurate reads that can differentiate between non-identical copies. In this simple model, the y-axis shows the expected NG50 contig size when repeats of a certain length (x-axis) or sequence identity (colored lines) can be consistently resolved. The “30x ultra” data point is a prediction, all others are real assemblies.
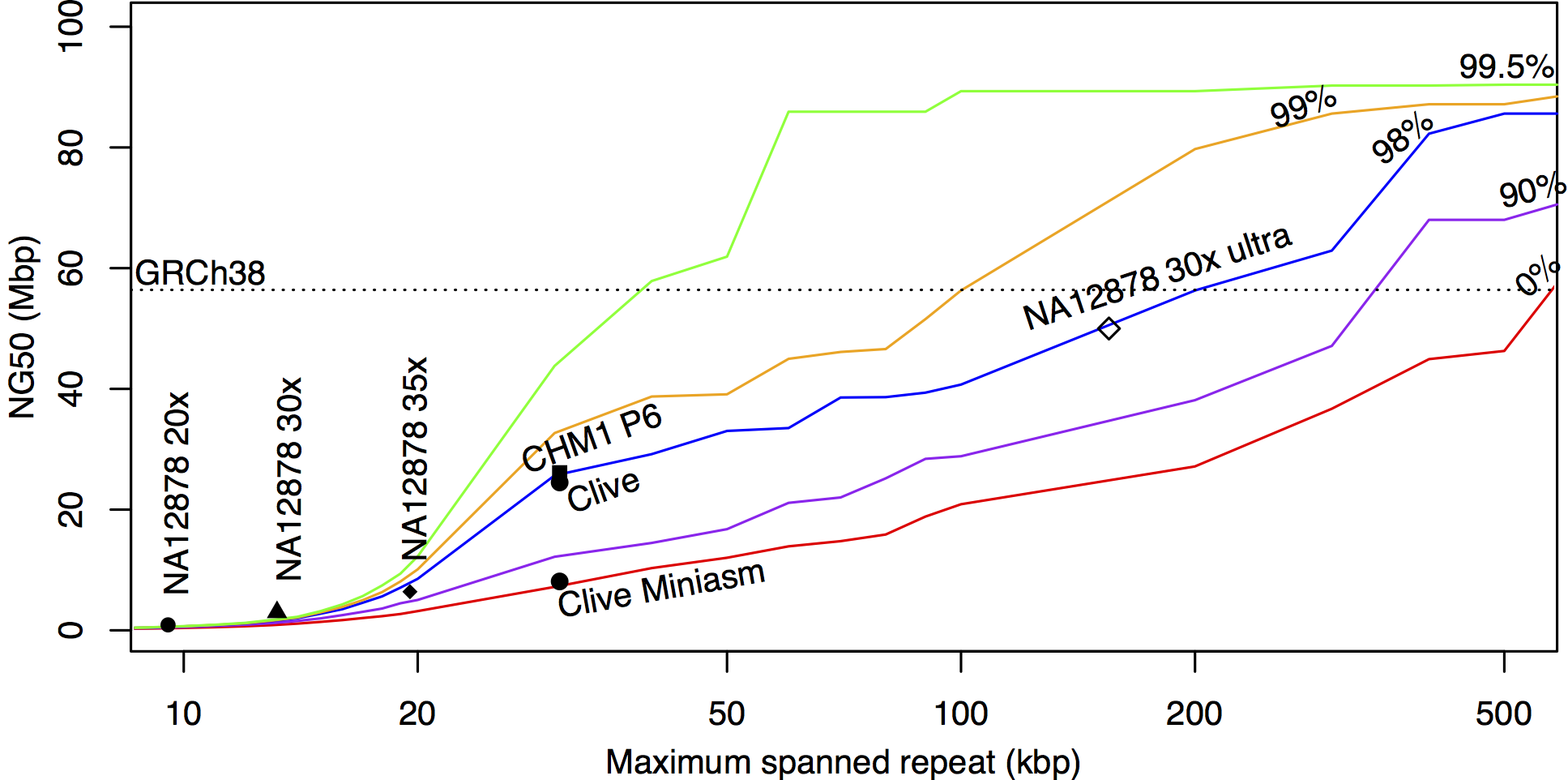
The dashed line shows the NG50 of the current human reference (GRCh38). Note that even for long read lengths, resolution of near-identical repeats is critical for a continuous assembly. For example, if reads are long enough to span 100 kbp repeats, but too noisy to differentiate between 90% identical repeats (purple line), contig NG50 is expected around ~30 Mbp. However, if those same reads were accurate enough to distinguish between 99% identical repeats (yellow line), continuity would be comparable to the human reference genome. Thus, assembly continuity depends on read length, accuracy, and coverage. Based on past experience, Canu can differentiate between ~98% identical repeats with PacBio or Nanopore (blue line). Good coverage is also very important for read correction, repeat spanning, and consensus accuracy; and increases in coverage will also increase continuity.
By this prediction, even without the ultra-long protocol, a standard 50x coverage nanopore dataset should exceed 20 Mbp NG50. Oxford Nanopore CTO Clive Brown happens to have sequenced his own genome to 55x coverage (Cliveome). [Edit 04/28/2017] As requested by Matt Loose, we computed the N50 of this dataset is 15.5 kbp and the average read length is 8.5 kbp. The dataset is copyrighted in such a way that we cannot re-distribute or publish it, but we have received Clive’s permission to report on our assembly. Data points for both Canu and miniasm assemblies have been added to the above figure.
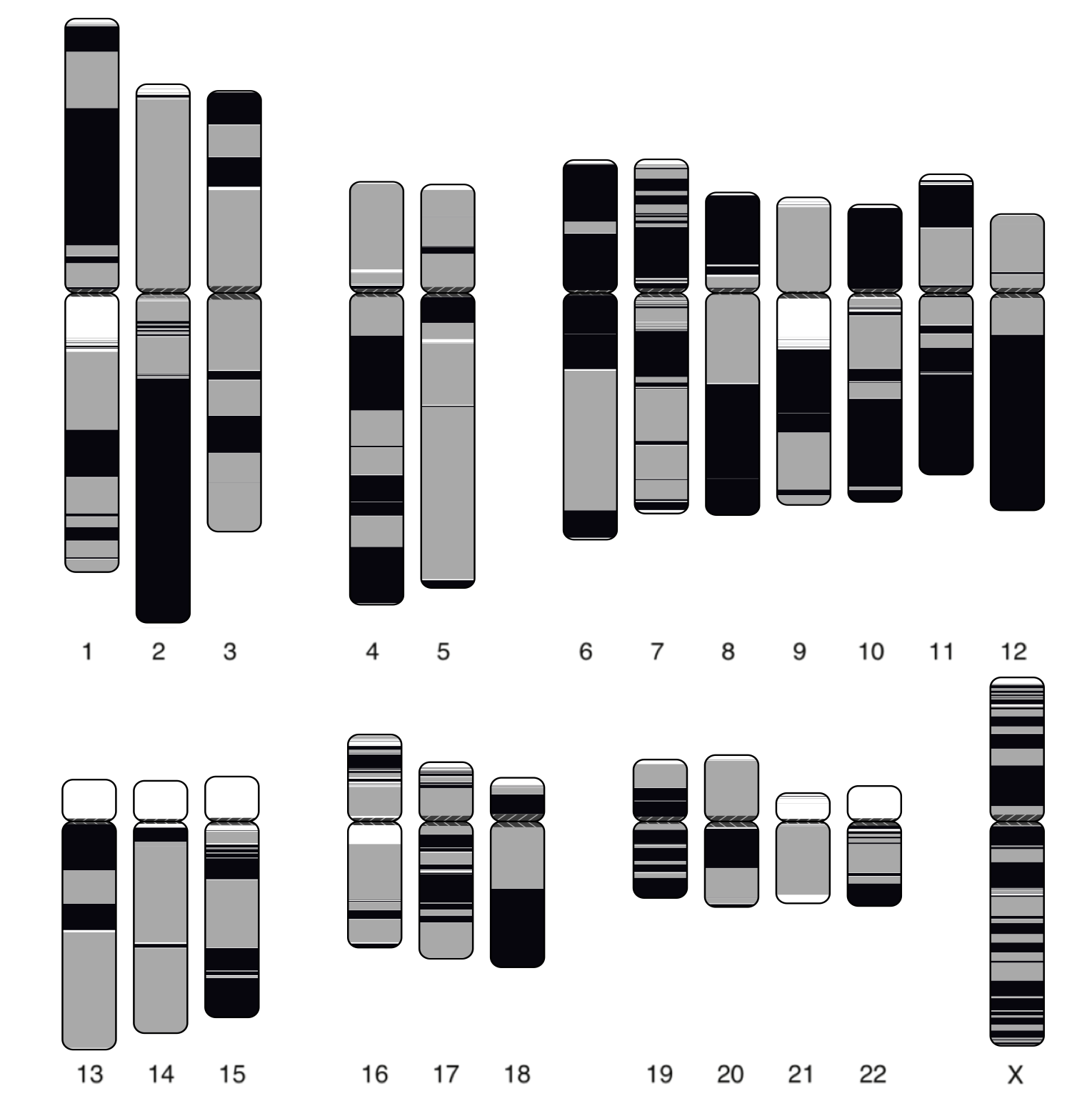
The Canu Cliveome assembly matches the model’s prediction, and falls right next to a recent 65x PacBio assembly of CHM1 in terms of continuity (Schneider et al. 2017). The Cliveome assembly comprises just 764 contigs, a maximum contig size of 109 Mbp, and a NG50 of 24.5 Mbp. This is one of the most continuous diploid human assemblies ever generated. Looking at an ideogram, most of the chromosomes are in less than 10 contigs:
Cliveome contigs overlaid on the human reference. Coloring (gray and black) is alternated for each alignment block, so continuous blocks indicate continuously mapping sequence and alignment or contig breaks appear as color switches. White indicates no coverage, usually corresponding to reference gaps (N’s).
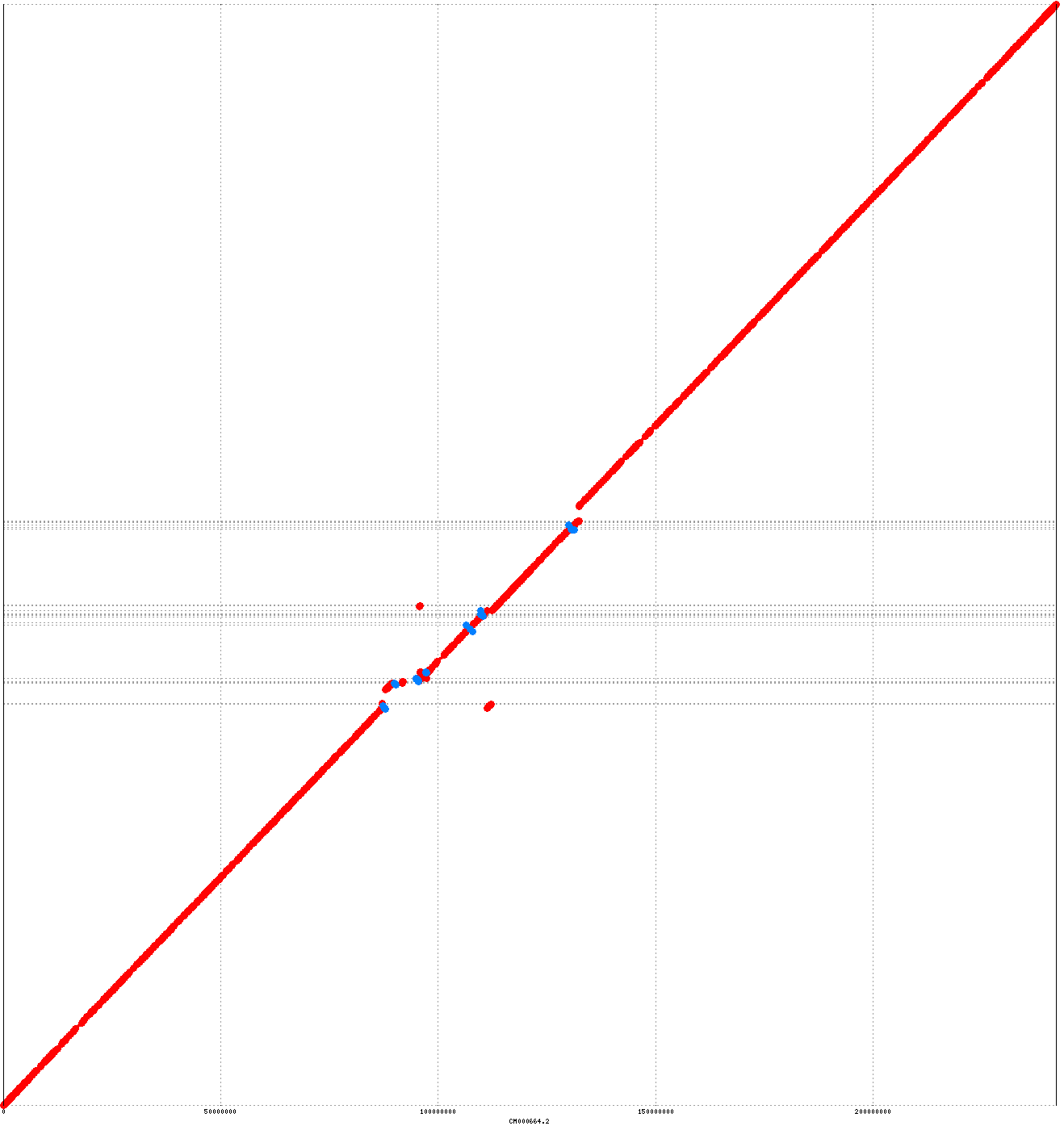
As reported by Nucmer, the assembly has 907 major structural differences from the reference, on par with previous PacBio human assemblies. [Edit 04/28/2017] In response to Jason Huff’s question regarding completeness/genes, we analyzed this genome using the GRCh38 reference. It covers 86.56% of 3.2 Gbp in the reference which includes alts. The CHM1 assembly in the figure above covers 86.73% of the same reference so the two are comparable in their completeness. We did not run LAP/CGAL since we did not have an independent short-read sequencing library of Clive available. An example nanopore assembly alignment is shown below for chromosome 2, where 78% of reference chromosome is contained in just two contigs, which both structurally agree with the GRCh38 reference:
Canu Cliveome assembly versus GRCh38 chromosome 2. Red matches on the main diagnoal indicate agreement, and off-diagnoal or blue matches indicate chimeric or inverted sequence (respectively). The chromosome arms are assembled in two large contigs, each over 80 Mbp in length. The centromeric region (middle of the figure) is fragmented and poorly aligned due to both the incompleteness of the reference and elevated repeat complexity.
Some of the differences between the assembly and the reference likely represent true variations between Clive’s genome and the reference. However, the assembly does appear to contain tens of large-scale errors, which would need correcting/validation with complementary scaffolding data. Consensus base accuracy is ~96.5%, similar to our unpolished assembly of GM12878. At the moment, base accuracy remains the biggest limitation of nanopore sequencing. As reported in Jain et al., moving to newer base-callers and adding signal-level polishing can improve accuracy to ~99%, but this does not yet match the >99.99% accuracy (QV40) achievable with current PacBio sequencing. In terms Keith Robison would prefer, that’s a difference between an error every 100 bases versus an error every 10,000 bases. That’s a serious difference, although, we expect continued improvements in nanopore consensus accuracy from new chemistries and base-calling algorithms.
Conclusion
The main takeaway is that nanopore sequencing can produce extremely continuous human assemblies, matching or exceeding the best PacBio assemblies, albeit with a sacrifice in accuracy. A second takeaway is the importance of repeat separation for assembly continuity. Even with ultra-long reads, there is a large difference between an assembly which resolves 85% identical repeats and one that resolves 98% identical repeats. Canu assemblies approximately follow the 98% line. In contrast, miniasm follows the 85% line, suggesting that it is not resolving closely-related repeats. This is likely due to Canu’s correction algorithm, which reduces the effective read error rate, allowing similar repeats to be distinguished. Nevertheless, miniasm is two orders of magnitude faster than Canu, and can be very useful to quickly triage and evaluate datasets (a common use case for our lab). Ideally, one could combine the speed of miniasm with Canu’s improved repeat separation. Canu’s correction stage is already quite fast (<10% of total runtime), so we are looking at speeding up the remaining stages and/or combining Canu read correction with other assemblers as was recently done for the tomato genome (Schmidt et al. 2017). Finally, we emphasize that de novo assembly may not be the best strategy for genotyping many human genomes. Because high-quality human reference genomes already exist, a hybrid strategy that combines reference (graph) mapping and localized de novo assembly may be best. Regardless, we are entering a new era of human sequencing and it will be exiting to watch how it sorts out.
You can find a description of our nanopore assembly methods in either the Jain et al. 2017 paper or our prior post on assembling GM12878 Assembly of a human genome from nanopore sequencing data. Sergey will be presenting this work at London Calling next week, and Adam will be presenting the week following at a Biology of Genomes workshop.