Assembly of a human genome from nanopore sequencing data
An international consortium recently released ~30x coverage of a human immortalized cell line (NA12878) sequenced using Oxford Nanopore MinION instruments. Release 3 of this dataset included 39 flowcells, which generated 14,183,584 reads and 91,240,120,433 bases, mostly using the 1D ligation prep, but with a few rapid kit runs as well. Our friends Nick Loman and Jared Simpson asked if we could assemble this data with Canu. Of course we said yes.
We had just released Canu v1.4 and were eager to try it on a large Nanopore dataset. In the Canu preprint (Koren et al. 2016) we report the successful assembly of a few bacteria and yeast. In these cases the Nanopore data assembled similarly to PacBio data, albeit at a lower consensus accuracy. From this experience, we expected the human assembly to roughly match what we see for PacBio data in terms of continuity, which can reach tens of mega-bases with good coverage.
All Nanopore data and the assembly described below are available on the NA12878 consortium GitHub page.
(EDIT:2017-01-09) Check out Mark Chaisson’s analysis of SVs in Clive Brown’s data for more insight on current Nanopore error modes: “SV Calling in the cliveome”
Continuity
As always, good assemblies are all about read length and coverage. We ended up doing both a 20x and 30x assembly because we launched the first one before all the data was available. Adding that additional 10x coverage made a big difference since this data included the rapid kit runs, which included the longest reads (some more 100 kb). Although the average read length did not change significantly between the two read sets (6.5 kb for both), the coverage in reads >10 kb increased from 10x to 16x. This resulted in a respectable contig NG50 of 3 Mbp for the 30x assembly. Here are the essential stats, where ‘NG50’ gives the contig size such that at least half of the 3.1 Gbp human genome is assembled into contigs of this size or larger:
20x assembly
Total units: 6297
BasesInFasta: 2590926517
Min: 1,779
Max: 9,317,548
NG25: 1,993,705 COUNT: 257
NG50: 887,678 COUNT: 835
NG75: 202,744 COUNT: 2561
30x assembly
Total units: 2886
BasesInFasta: 2646010004
Min: 1,673
Max: 27,160,256
NG25: 6,437,016 COUNT: 80
NG50: 2,963,950 COUNT: 266
NG75: 670,702 COUNT: 776
Illustrating the importance of read length, an older 50x PacBio NA12878 dataset with an average read length of 4.5 kb assembled with an NG50 of 0.9 Mbp (Pendleton et al. 2015), a third of this assembly. In comparison, newer PacBio human assemblies, with average read lengths of >10 kb, can reach NG50 values of more than 20 Mbp (Schneider et al. 2016). Judging from these early results, and given higher coverage and longer libraries, we expect future Nanopore assemblies will reach similar contig sizes.
Accuracy
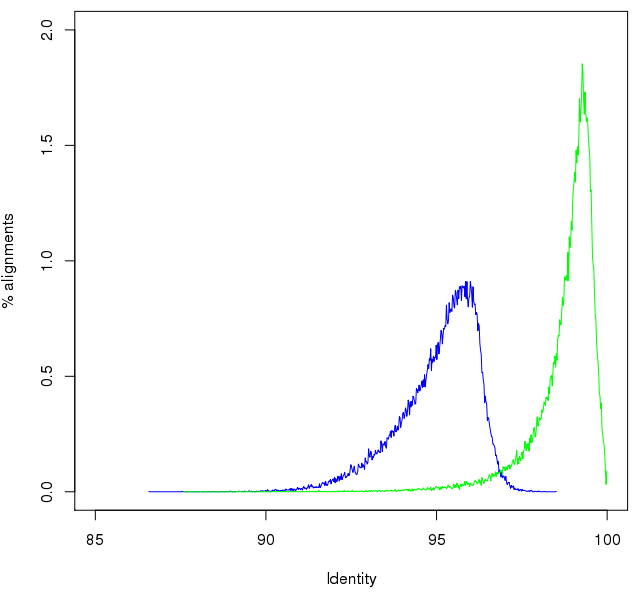
Canu uses a hierarchical strategy to correct reads before assembly. Due to what appears to be systematic error in the Nanopore base calls, our corrected reads were were less accurate than usual. While corrected PacBio reads are typically >99% identity, these Nanopore reads averaged only 92% after correction. Similarly, the average consensus accuracy of the assembled contigs was only 95%, measured against the GRCh38 reference. Using the raw reads, we tested an alternate consensus tool Racon (Vaser et al. 2016) on a subset of contigs, but consensus accuracy increased just 1%, suggesting an issue with the reads themselves rather than the Canu algorithm. The real gain comes from polishing the Nanopore assembly with Illumina data, which boosts consensus accuracy up to 99%.
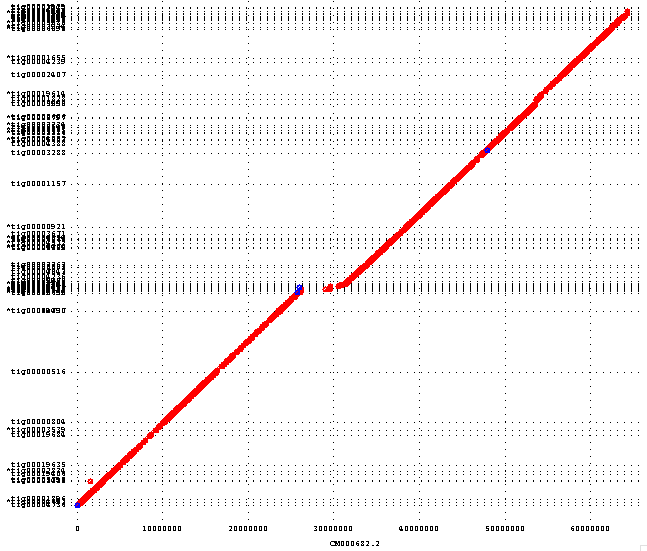
We are in the process of evaluating the structural accuracy of this assembly versus GRCh38, but initial results look good and the assembled contigs map well to the reference. In particular, the Nanopore reads seem to have a lower rate of chimerism than PacBio reads. This improves the throughput of the correction process (i.e. fewer reads are broken during trimming) and reduces the risk of a chimeric read falsely joining two contigs. The following dotplot shows GRCh38 chromosome 20 along the X axis and assembled contigs laid out along the Y axis. Each horizontal gridline represents a contig boundary. The jump in the middle is the centromere, which is incomplete in the reference and cannot be mapped:
Runtime
Canu was about fourfold slower on the Nanopore data compared to similar coverage of PacBio, requiring ~60,000 CPU hours for the 30x assembly. To some extent, this is not surprising due to the higher noise of the input reads. However, systematic error in the Nanopore reads also caused our overlapping step, which uses k-mers to estimate overlap quality, to explore more candidate overlaps than usual. Tweaking the overlapping parameters helped address this issue. The systematic error also increased the residual error of the corrected reads, which increased runtime of the downstream steps.
Super-long reads (>100 kb) are another reason for the increased runtime. For corrected reads, Canu still uses old Celera Assembler alignment code that was originally designed for Sanger sequencing data. Unlike the MinHash method we currently use for raw read overlapping, these older methods extend alignments from single k-mer seeds. This works OK for corrected PacBio reads, which are 10’s of kb and highly accurate, but is inefficient for the much longer (and noisier) Nanopore data. We are working to replace these steps with the same MinHash approach used in the earlier stages.
Conclusion
Overall this first Nanopore human assembly was a success. The continuity is that (or better) of a similar coverage PacBio run using current chemistries. The encountered issues boil down to systematic base-call error and inefficiencies caused by the super-long read lengths. We are testing improvements for the latter, but it is up to Nanopore to fix the former. Notably, the consensus accuracy we see here is significantly lower than for bacterial genomes like E. coli, which can reach 99%. This suggests that the Nanopore base caller is underperforming on human, either due to DNA modifications or sequence contexts not seen in the training data. We are also testing improvements to the Canu correction module to make better use of the data we have. Until these issues can be resolved, polishing Nanopore assemblies with Illumina data can improve accuracy in the unique regions of the genome. However, this approach does leave some sequence uncorrected because the Illumina reads cannot be uniquely mapped to the entire genome. This is the main limitation of Nanopore assembly at this time, compared to PacBio, which can produce high base accuracy across the entire genome.
Computational details
20x assembly
An initial data release contained 20x coverage which we assembled as a trial run using Canu v1.4 (+11 commits) r8006 4a7090bd17c914f5c21bacbebf4add163e492d54:
canu -p asm -d asm genomeSize=3.1g gridOptionsJobName=na12878nano "gridOptions=-t 72:00:00 -p norm" -nanopore-raw rel2*.fastq.gz corMinCoverage=0 corMaxEvidenceErate=0.22 errorRate=0.045
These are our standard low-coverage parameters, but with a decreased max evidence error rate to reduce memory requirements. From previous experience, we suspected our MinHash overlapping algorithm was over-estimating overlap identities due to systematic error in the base calls. (Counterintuitively, this systematic error makes two reads look more similar than reality due to the fact that they share more k-mers than expected under a random model). Manually decreasing the maximum overlap error rate used for correction adjusted for this bias. The assembly took 40K CPU hours (25K to correct and 15K to assemble). This is about twofold slower than a comparable PacBio dataset.
30x assembly
Canu v1.4 (+11 commits) r8006 4a7090bd17c914f5c21bacbebf4add163e492d54:
canu -p asm -d asm genomeSize=3.1g gridOptionsJobName=na12878nano "gridOptions=-t 72:00:00 -p norm" -nanopore-raw rel3*.fastq.gz corMinCoverage=0 corMaxEvidenceErate=0.22 errorRate=0.045
This initial correction took over twice as long as the initial run (>50K cpu hours). Again we suspected the base call bias was to blame, so we tweaked the overlapping parameters to decrease the number of hashes used and increase the minimum identity threshold. This reduces the sensitivity of the algorithm to improve runtime:
canu -p asm -d asm genomeSize=3.1g gridOptionsJobName=na12878nano "gridOptions=-t 72:00:00 -p norm" -nanopore-raw rel3*.fastq.gz corMinCoverage=0 corMaxEvidenceErate=0.22 errorRate=0.045 "corMhapOptions=--threshold 0.8 --num-hashes 512 --ordered-sketch-size 1000 --ordered-kmer-size 14"
This assembly required 62K CPU hours (29K to correct, 33K to assemble), which is about fourfold slower than a comparable PacBio dataset.
Comparison to the reference
The 30x assembly was aligned to the GRCh38 reference using MUMmer:
nucmer -l 10 -c 1000 hg38.fna asm.contigs.fasta
dnadiff -d out.delta
mummerplot --large --fat --png out.1delta
The average identity to the reference was 95.34%. (Note: these alignment parameters are not ideal and we plan to redo the comparison). After Pilon polishing with Illumina data, the assembly was aligned again:
nucmer -l 10 -c 1000 hg38.fna asm.polished.fasta
dnadiff -d out.delta
mummerplot --large --fat --png out.1delta
Which increased the identity to 99.02%. However, there is a long tail of unpolished sequence remaining (5% of total) at less than 98% identity. The figure shows the initial (blue) and polished (green) alignment identities: