The Q100 preprint!
We are delighted to finally announce a preprint describing the Q100 project, where we finished the HG002 genome to near-perfect accuracy: A complete diploid human genome benchmark for personalized genomics
Building benchmarks is hard, unglamorous work, but the impact can be huge. Consider how much the Genome in a Bottle (GIAB) variant benchmarks have shaped the field over the past ~10 years. However, these mapping-based benchmarks omit about 15% of the genome (and not just in centromeres). Assembling and annotating the complete, diploid HG002 genome from “T2T” allowed us to fill in those missing regions and explore the limitations of reference-based variant calling and benchmarking, especially within complex, segmentally duplicated regions that are often heterozygous.
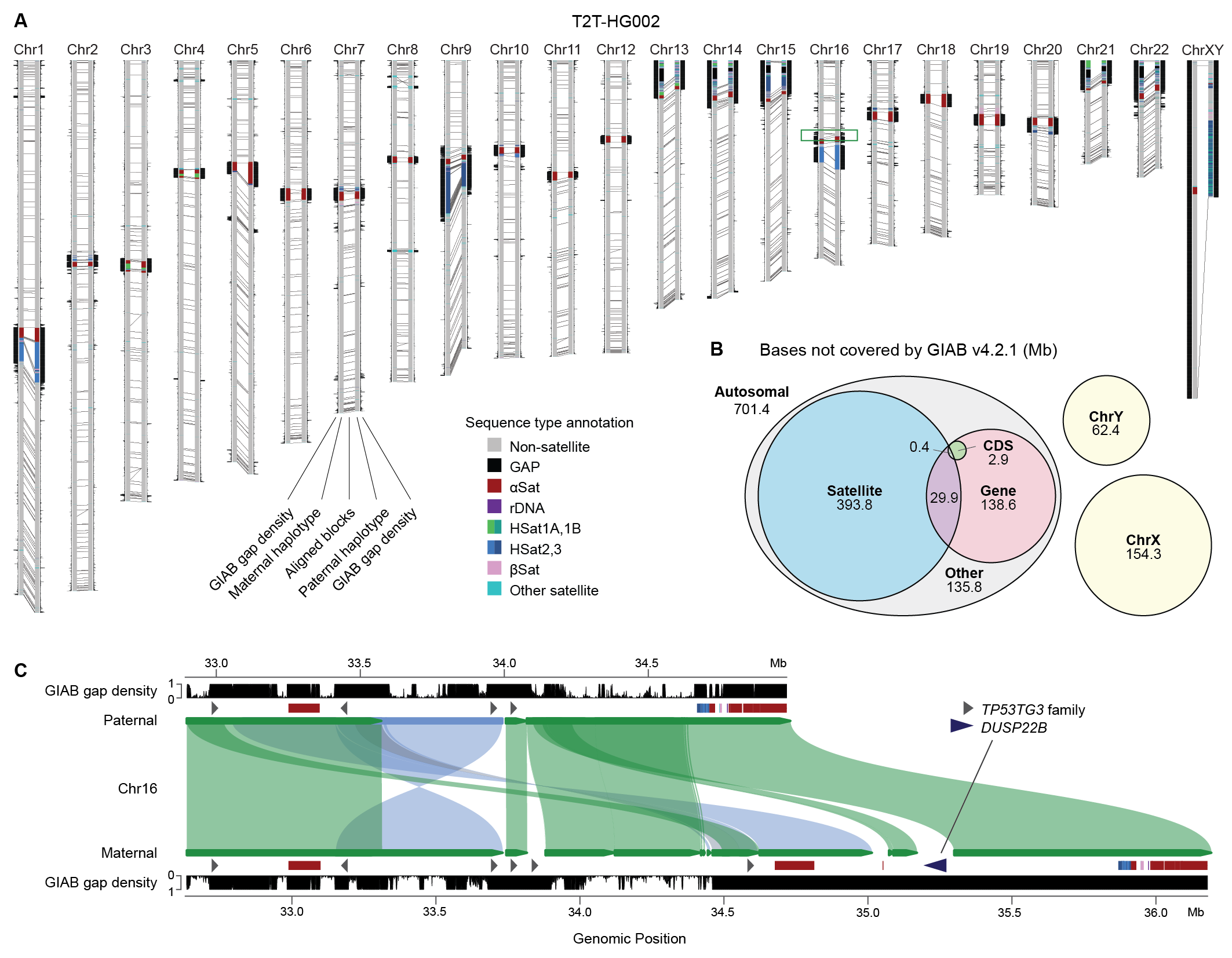
In our usual fashion, we ran this project entirely in the open, with the first T2T-HG002 assembly released on GitHub back in Nov 2022. What took so long to write the paper? We spent a lot of time checking our work, but we also had to develop new methods for benchmarking against a complete, diploid T2T genome. Enter Genome Quality Checker (GQC) by project leader, Nancy Hansen. With a complete benchmark and appropriate QC methods now in place, we can measure the accuracy of assemblies, variant callsets, and even raw reads across the entire genome.
This revealed something surprising: long-read de novo assembly methods now outperform reference-based variant calling not just in completeness, but also in overall accuracy, by a substantial margin (10 QV)! Most of this gain comes from “hard to call” regions of the genome, but the result still holds when looking only at regions where the variant caller (e.g. DeepVariant) reports high confidence (GQ >40). We are actively digging into this result, but it hints at plenty of room for improvement in modern variant calling methods beyond the 0.999 F1 scores we’ve come to expect from variant benchmarks.
A huge thanks to Nancy Hansen and the whole Q100 team for shepherding this project over the past 3 years. Along the way, we made new friends who used our methods (e.g. Verkko, Merqury) to assemble near-perfect T2T genomes of their own, including an East Asian diploid reference, a South Asian diploid reference, a rhesus macaque reference, and complete diploid assemblies of the human cell lines RPE-1 and BJ and IMR-90. Each of these studies includes their own unique twists and insights, and we encourage you to read them as well.
Clinically routine T2T genomes are finally in sight!