The (near) complete sequence of a human genome
The Telomere-to-Telomere (T2T) consortium is proud to announce our v1.0 assembly of a complete human genome. This post briefly summarizes our work over the past year, including a month-long virtual workshop in June, as we strove to complete as many human chromosomes as possible. Our progress over the summer exceeded our wildest expectations and resulted in the completion of all human chromosomes, with the only exception being the 5 rDNA arrays. Our v1.0 assembly includes more than 100 Mbp of novel sequence compared to GRCh38, achieves near-perfect sequence accuracy, and unlocks the most complex regions of the genome to functional study. We plan to release a series of preprints in the coming months that fully describe our methods and analyses, but due to its tremendous value, we are releasing the assembly immediately.
Roughly twenty years ago, the International Human Genome Sequencing Consortium published an “Initial Sequencing and Analysis of the Human Genome” simultaneously with “The Sequence of the Human Genome” from Celera Genomics. Although the public consortium chose a more humble title that suggested some work was left to be done, amidst all the pomp it was easy to miss the fact that the human genome had not actually been finished. The key caveat appears early on in both papers, e.g. “A 2.91-billion base pair (bp) consensus sequence of the euchromatic portion of the human genome was generated”. Ignoring heterochromatin, due to difficulty in mapping, cloning, or assembling these sequences, excluded upwards of 10% of the genome from these initial drafts, and that missing fraction has been underappreciated ever since. Today, the latest human genome reference (GRCh38) still contains 161 Mbp of “unknown” sequence constituting 5% of the genome.
Now, twenty years later, we are finally able to fill in the blanks thanks to a confluence of new sequencing technologies from PacBio and Oxford Nanopore. Within the past year, the T2T consortium assembled the first complete human chromosomes, Chromosome X and Chromosome 8, using Nanopore ultra-long (UL) sequencing as a backbone and polishing that sequence with PacBio and Illumina. However, the recent release of PacBio’s HiFi technology led us to revise our recipe. In the assembly presented here, we first constructed a highly-accurate assembly graph using PacBio HiFi reads and then resolved any structural ambiguities with the help of Nanopore UL reads. The following image shows a Bandage visualization of our HiFi string graph for the CHM13 genome, with most chromosomes resolved as individual components.

After resolution of the remaining “tangles” with the help of Nanopore UL reads, the sequence of each complete chromosome was obtained via a consensus of HiFi reads taken from the corresponding traversal of the graph. This approach allowed us to reach T2T continuity for all chromosomes, while retaining the accuracy of PacBio HiFi throughout. Nanopore data also helped by patching a few regions of the genome that PacBio failed to sequence due to an apparent sequencing bias in GA-rich repeats. Similar graph-based hybrid approaches have previously been used to combine Illumina and long-read sequencing data for microbial genomes, but this has not been possible for human genomes due to the larger genome size and higher repeat complexity. However, because HiFi reads are both long and accurate, the complexity of the resulting HiFi graph is tremendously reduced compared to Illumina, allowing for complete resolution of the remaining tangles via Hamiltonian paths and, in the worst cases, Nanopore threading.
We estimate that the consensus quality of our HiFi-based assembly exceeds Q60 (less than 1 error per million bases), with most remaining errors localized to homopolymers, which is a known issue for both PacBio and Nanopore sequencing. To correct homopolymer errors and further improve quality, we mapped the raw PacBio, Nanopore, and Illumina reads to our initial assembly and called variants using DeepVariant and Sniffles. Considering only the most confident variant calls (likely to be assembly errors, rather than heterozygosity), we made a total of 4 structural corrections and 993 small variant corrections. We estimate that the quality of our polished assembly approaches Q70 (1 error per 10 million bases), with no known structural errors. We plan to continue scrutinizing the assembly over the coming months and will generate updated versions to correct any errors identified.
Both the v0.9 pre-polished and v1.0 polished assembly are now freely available for download. The CHM13 cell line possesses a 46,XX karyotype with almost complete homozygosity between chromosome pairs. As such, the v1.0 assembly contains 23 chromosomes (no ChrY) and 1 mitochondrial genome totaling 3,045,441,522 bp of assembled sequence. A small number of heterozygous variants were observed in the genome and will be fully cataloged in a future release. As mentioned above, only the 5 rDNA arrays remain unfinished. Some full and partial rDNA units are assembled on each of the 5 acrocentric p-arms, but the centers of these arrays are currently represented by a total of 11.5 Mbp of Ns in the assembly (on Chromosomes 13, 14, 15, 21, 22). Because the rDNA arrays are near-identical tandem repeats, the content of these arrays is known and only the contained variants remain to be determined. We expect to finish these arrays and add a Chromosome Y (from a different cell line) in the coming year.
Due to our rapid release timeline, many details of our methods and validation have been omitted here. We plan to post a preprint fully describing this project in the coming months, and our consortium is in the process of characterizing these newly uncovered regions of the genome. We have freely released all of our raw data and assemblies without restriction, but ask as a courtesy that you contact us if you would like to contribute analyses prior to our initial publication of results in approximately 6 months. The T2T is an open consortium and all are welcome to join our effort to generate the first truly complete assembly of a human genome. If you would like to join us, please contact T2T co-chairs Adam Phillippy adam.phillippy@nih.gov and Karen Miga khmiga@ucsc.edu to be added to our mailing list.
Acknowledgements
Who is the T2T consortium? You can find a continually updated list of contributors on our members page. I would like to sincerely thank everyone involved for their dedication over these past few months. An incredible amount of work was accomplished in a very short time. Thanks also to our industry partners at PacBio, Oxford Nanopore, Arima Genomics, Amazon Web Services, and DNAnexus who helped enable this work.
I would especially like to thank the working group co-chairs who helped me organize this summer’s finishing workshop: Sergey Nurk (assembly), Karen Miga (satellite DNAs), Arang Rhie (polishing and validation), Mark Diekhans (browser and annotation), Mitchell Vollger (segmental duplications), and Justin Zook (variant calling). Sergey Nurk deserves special credit for developing the HiFi assembly methods that enabled such rapid progress this year.
Lastly, I would like to acknowledge an abbreviated list of software that was critical to the success of this project: HiCanu, Miniasm, Winnowmap, Minimap, GraphAligner, MUMmer, CentroFlye, TandemTools, StringDecomposer, Bandage, GFA, IGV, DeepVariant, Merqury, and SDA. In particular, the contributions of Heng Li in terms of both tools and formats have been invaluable. Never forget that the field of genomics is entirely dependent on free software and the developers of these tools deserve endless thanks and support!
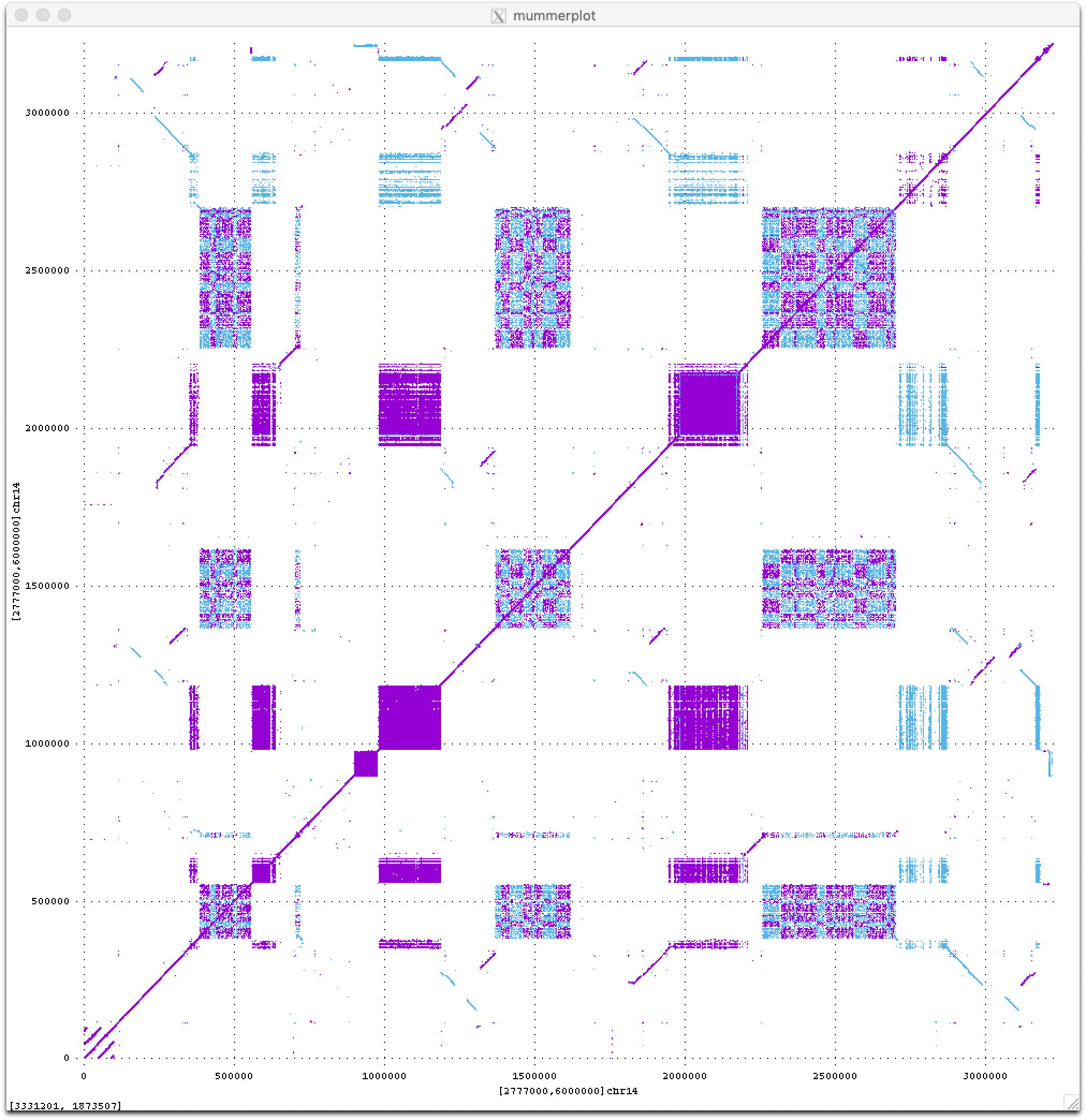
Stay tuned for much more to come from the T2T consortium this year as we begin to analyze these new sequences of the human genome. For now I will just leave you with a teaser dot plot of all ≥50 bp exact repeats within a novel 3 Mbp region on the short arm of human Chromosome 14, which I think looks quite beautiful (fwd in purple, revcomp in blue).